5. Simulation of a PSI 1,3,5-trimethylbenzene Experiment
Task 5.1 Simulation of 1,3,5-trimethylbenzene experiment
The previous tasks have focused on using AtChem to carry out detailed gas-phase photochemical chamber box modelling of experiments conducted at the outdoor EUropean PHOto-REactor (EUPHORE) in Valencia. Here we look at constructing a chamber optimised AtChem box model, incorporating the MCMv3.1 degradation scheme for the model anthropogenic aromatic compound 1,3,5-trimethylbenzene (TMB), in order to simulate data measured during an experiment carried out in the Paul Scherrer Institut (PSI) aerosol chamber facility in Switzerland (Paulsen et al. (2005)).
During the experiment discussed below, comprehensive high (mass and time) resolution measurements of TMB and its photo-oxidation products were recorded by a novel Chemical Ionisation Reaction Time-of-Flight Mass Spectrometer (CIR-TOF-MS). The detailed box model can be used to interpret this dataset in detail. Model-measurement comparisons, using such highly detailed simultaneous measurements of the complex array of multi-functional products formed, enables us to evaluate the MCM mechanism in detail and can also provide key insight for guiding the directions of future laboratory experiments. For more information see Rickard et al. (2009) and Wyche et al. (2009).
5.1 Model Mechanism
Locate the TMB mechanism on the MCM website (MCM name 'TM135B'). Extract this mechanism along with the set of inorganic reactions and save as an appropriate AtChem mechanism file (extension '.fac').
5.2 Dilution
As for any chamber, dilution of stable reactants and products (via replenishment of gases sampled or lost from the chamber) needs to be taken into account. You can use the VARIABLES output from the extracted MCM TMB mechanism to define the dilution reactions of the stable reactants and products. However, to save time, a set of dilution reactions of stable species in the TMB mechanism is listed in the file tmb_dilution.fac. Copy these reactions and append to the end of the mechanism in your mechanism file.
5.3 PSI Chamber Auxiliary Mechanism
As with the EUPHORE chamber, the 'background' chamber chemistry needs to be taken into account. The chamber specific auxiliary mechanism employed here for the PSI chamber was optimised in separate characterisation experiments designed specifically to investigate how the chamber surface reactions vary temporally over a range of conditions. From sensitive measurements of HONO in the photooxidation of TMB, Metzger et al. (2008) found that the addition of a parameterised (light-induced) heterogeneous NO2/HONO conversion reaction in the chamber specific auxiliary mechanism significantly improved model performance under a variety of VOC/NOx conditions.
The optimised PSI auxiliary chamber mechanism as derived in the experiments of Metzger et al. . (2008) is given in psi_chamber.fac. The off-gassing of organics from the wall that convert OH to RO2/HO2 leads to rapid ozone formation during pure air irradiations. A dummy reaction of X + OH -> X + HO2 is included in the auxiliary mechanism to account for this with a nominal [X] = 300 ppbv assumed for all TMB experiments (see Table 7). For more information see Metzger et al. (2008). Open psi_chamber.fac, copy the mechanism and append to the end of your mechanism in your mechanism file (after the dilution reactions).
5.4 Photolysis Rate Parameterisations and Fixed Concentration Species
Now that the mechanism file has been defined for use in modelling TMB experiments in the PSI chamber we now have to set up the photolysis rates and other Environmental Constraints.
The photolysis rates in AtChem are currently set up for use with the outdoor EUPHORE chamber. The PSI aerosol chamber is a temperature controlled indoor chamber comprising a 27 m3 Teflon bag illuminated by four 4 kW xenon arc lamps (Paulsen et al., 2005). Measurements of all important photolysis rates have been made in the PSI chamber and the photolysis rate parameterisations have been tuned accordingly (Paulsen et al., (2005); Metzger et al., (2008)). To make life simple, these PSI photolysis rate parameterisations, for all photolysis reactions, have been calculated and are given in the file photolysis_rates.const, save this file to your computer. Note the extension of '.const' of this photolysis file, this lets AtChem know that the photolysis parameterisation is of a different format which gives a constant photolysis rate for the duration of the experiment. This file should be uploaded to the photolysis rates input box.
The dummy species 'X', as defined in the auxiliary mechanism and Table 7 also needs to be set to a constant 300 ppbv throughout the experiment. This is be done by entering the species name (X) and the concentration (in molecules cm-3) into the fixed-concentration species box near the bottom of the AtChem input page. The species name and concentration need to be separated by a space, more than 1 species can be fixed in this way and each species should be entered on a seperate line.
5.5 Set up the Initial Concentrations File
The initial conditions of the PSI TMB experiment to be modelled are given below in Table 7.Set up an initial concentration file (using the extension '.config') in order to model the PSI TMB experiment
| 27/11/2006 | |
|---|---|
| Start time (hh:mm) | 08:00 |
| End time (hh:mm) | 18:00 |
| TMB (ppbv) | 597.0 |
| NO (ppbv) | 135.0 |
| NO2 (ppbv) | 130.0 |
| H2O (molecules cm-3) | 2.89 x 10+17 (or 50 % RH) |
| Dilution Rate (s-1) | 8.64 x 10-6 |
| Taverage (°C) | 20 |
| X (ppbv) | 300 |
5.6 Set up the Concentrations Output File
The object of this exercise is to model the TMB photo-smog experiment carried out on the 271106 and to compare simulations of the gas-phase evolution of various aspects of the TMB system with detailed CIR-TOF-MS measurements in order to gain insights into the complex chemistry occurring. Figure 2 highlights the major primary and secondary product formation routes in the OH initiated photooxidation of TMB mechanism in MCMv3.1. As shown in Figure 2 there are numerous ring opening and ring retaining products formed.
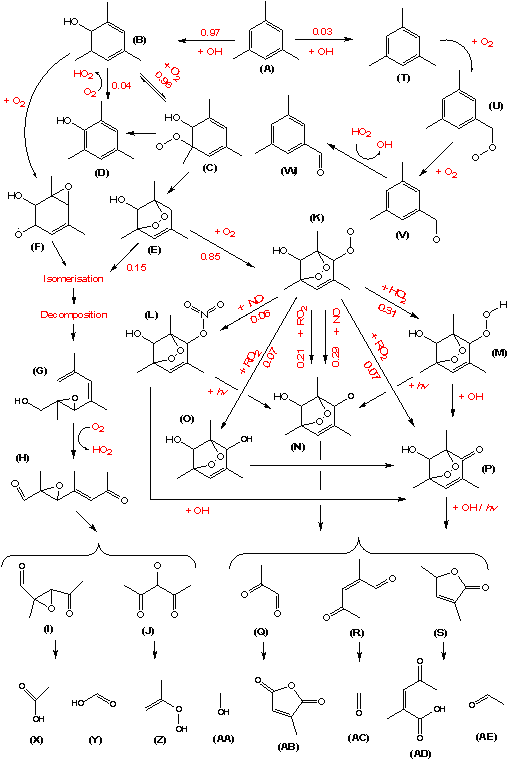 |
| Figure 2. Summary of OH radical-induced photooxidation of 1,3,5-trimethylbenzene. Key primary and secondary product formation shown with branching ratios from the MCMv3.1 (after Wyche et al., (2009)) |
Construct a concentration output file (with the '.config' extension) with the following species included (letters in brackets refer to structures in Figure 2):
TM135B, O3, NO, NO2, OH, HO2
Peroxide bicyclic ring opening products:
methyl glyoxal (Q)
3,5-methyl-5(2H)-2-furanone ((S), MXYFUONE)
2-methyl-4-oxo-2-pentanal ((R), C5MDICARB)
3-methyl maleic anhydride ((AB), MMALANHY)
Peroxide bicyclic ring-retaining products:
Hydroxyl peroxide bicyclic nitrate ((L)),TM135BPNO3)
hydroxy peroxide bicyclic ketone ((P), TM135OBPOH)
hydroxy peroxide bicyclic diol ((O) TM135BP2OH)
hydroxy peroxide bicyclic peroxide ((M), TM135BPOOH).
5.7 Set up/running the TMB model on AtChem
Now we have set up the appropriate input and output files we can set up the AtChem model run:
- (a) Open the AtChem homepage and click the link to Run AtChem Online
- (b) Give the model run an appropriate name and you may add some specific information on the model run in the 'description' text box if you wish.
- (c) Upload the mechanism file, initial concentration file, concentration output file and the photolysis_rates.const file in the appropriate boxeli.
- (d) Set up the 'Environmental Variables':
- (e) Set up the 'Model Parameters':
- (f) Enter the fixed concentration species X as described above
- (g) Running the model on AtChem
Firstly you will need to define or calculate the third body concentration M (in molecules cm-3). You can either specify M at typical ambient atmospheric temperatures and pressures or you can calculate M by entering 'CALC' into the 'M' box and specifying the temperature and pressure (NB: if you enter a value for M then you do not need to specify a pressure). The experimental value of M in this case is ~ 2.51 x 10+19 molecules cm-3
Either the average Relative Humidity (RH) or water concentration needs to be set in the model. Here either set RH to 50 % and H2O to 'CALC' or use the H2O concentration value specified in Table 7 (setting RH to 'NOTUSED').
Use the default DEC value (we are not using it anyway here). The 'DILUTE' value should be set to that specified in Table 7, 'JFAC' and 'ROOFOPEN' variables are not used here as we are constraining the photolysis rates.
Here we need to set up the start time of the model, the number of step sizes (i.e. how frequently we want to output the model species concentrations) and the date and location (in order to calculate the appropriate photolysis rates).
We want to run the TMB model for 10 hours starting at 08:00. Set the 'model start time', 'number of steps' and the 'step size' so that the model to outputs the model species concentrations (as defined in the 'Concentration Output' file) every 5 minutes for 10 hours.
All other model parameters can be left as defaulted for the purpose of this exercise.
Now you have set up the model run, click on the 'Run' button at the bottom of the page
5.7 AtChem Model Output
Hopefully the model should have run without any problems and the 'Model Execution Output' page should have opened. Listed on this page are the model statistics and where you can download the model input and output files.
Download the model output '.zip' folder (and output '.zip' folder if you wish) and save it to an appropriate place.
Open this folder and look in the 'ModelOutput' folder. This folder contain many files detailing the output parameters of the model run. Unzip and open the 'concentration' output file. This should give the time and model concentrations of all species defined in the 'Concentration Output' file, output every 5 minutes for 10 hours.
5.8 Model-measurement Comparisons
(i) Ring Opening Products
Open the 'concentration' output file in an appropriate graphical spreadsheet application and plot the concentration-time profiles of the ring opening products methyl glyoxal (Q), 3,5-methyl-5(2H)-2-furanone ((S), MXYFUONE), 2-methyl-4-oxo-2-pentanal ((R), C5MDICARB) and 3-methyl maleic anhydride ((AB), MMALANHY).
Compare the model results for methyl glyoxal with the experimental data contained in psi_tmb.xls by pasting your results into the 'mgly' worksheet.
How do your modelled results and calibrated CIR-TOF-MS methyl glyoxal profiles compare?
What does this tell us about how good a job (or not) the MCMv3.1 TMB mechanism is doing in describing the chemistry that is going on under the conditions of the experiment being modelled?
MXYFUONE, C5MDICARB and MMALANHY all have the same mass (m/z = 112) and therefore the CIR-TOF-MS is only able to attributed a signal to the sum of these compounds (@ m/z = 112+1).
Compare your model results and the sum of modelled m/z = 112 ring opening products with the experimental data contained in psi_tmb.xls by pasting your results into the 'ringopen' worksheet.
How do your modelled 112 profiles and calibrated CIR-TOF-MS m/z = 113 profile compare?
(ii) Ring Retaining Products
Open the 'concentration' output file in an appropriate graphical spreadsheet application and plot the concentration-time profiles of the peroxide ring ring retaining products: hydroxyl peroxide bicyclic nitrate ((L)),TM135BPNO3), hydroxy peroxide bicyclic ketone ((P), TM135OBPOH), hydroxy peroxide bicyclic diol ((O) TM135BP2OH) and hydroxy peroxide bicyclic peroxide ((M), TM135BPOOH).
Compare the model results for TM135BPNO3 and TM135OBPOH with the CIR-TOF-MS time profiles contained in psi_tmb.xls by pasting your results into the 'ringret' worksheet.
NB: Such high mass, relatively stable species as these multi-functional peroxide bicyclics have not been previously observed in the gas-phase, therefore we can only really compare the temporal profiles of the measured and modelled data (measured concentrations are calculated using a proxy calibrant and are effectively arbitrary).
How do your modelled peroxide bicyclic and CIR-TOF-MS profiles compare. Do you think these ring retained species are actually formed in the photooxidation of TMB under the conditions of the PSI experiment?
(iii) Peroxide Formation
Evidence exists to show that organic hydroperoxides may play a significant role in secondary organic aerosol (SOA) formation and subsequent growth (for more information see Wyche et al. (2009) and Rickard et al. (2009)). In the modelled PSI TMB experiment the most dominant organic hydroperoxide available early on in the chamber matrix is the O2-bridged peroxide TM135BPOOH ((M) in Figure 2)). However, such peroxides are not easily measured using the PTR ionisation technique used by the CIR-TOF-MS, primarily owing to substantial post ionisation fragmentation.
Plot the simulated temporal evolution of the multi-functional peroxide species TM135BPOOH with the time profile of the measured SOA particle number density (cm-3) contained in psi_tmb.xls by pasting your results into the 'peroxide' worksheet.
Comment on whether or not this peroxide species could be involved in the formation of SOA in the TMB experiment being modelled.
Next
Previous
Return to Tutorial homepage